# Load necessary packages using pacman for easier dependency management
pacman::p_load(
tidyverse, # Collection of R packages for data science (ggplot2, dplyr, etc.)
showtext, # Enables custom fonts for ggplot2
ggtext, # Adds rich text formatting to ggplot2
skimr, # Provides summary statistics in a readable format
glue,
janitor,
gganimate,
ggrepel,
legendry
)
# Add Google fonts
font_add_google("Oswald", family = "Oswald")
font_add_google("Roboto Condensed", family = "Roboto")
# Add local font
font_add("Font Awesome 6 Brands", here::here("fonts/otfs/Font Awesome 6 Brands-Regular-400.otf"))
# Automatically enable the use of showtext for all plots
showtext_auto()
# Set DPI for high-resolution text rendering
showtext_opts(dpi = 300)How This Graphic Was Made
1. 📦 Load Packages & Setup
2. 📖 Read in the Data
3. 🕵️ Examine the Data
glimpse(who_tb_data)
skim(who_tb_data)4. 🤼 Wrangle Data
# A tibble: 24 × 18
country g_whoregion iso_numeric iso2 iso3 year c_cdr c_newinc_100k cfr
<chr> <chr> <dbl> <chr> <chr> <dbl> <dbl> <dbl> <dbl>
1 Democrat… South-East… 408 KP PRK 2000 28 144 0.31
2 Democrat… South-East… 408 KP PRK 2001 24 123 0.33
3 Democrat… South-East… 408 KP PRK 2002 33 168 0.3
4 Democrat… South-East… 408 KP PRK 2003 40 205 0.27
5 Democrat… South-East… 408 KP PRK 2004 36 184 0.28
6 Democrat… South-East… 408 KP PRK 2005 34 175 0.29
7 Democrat… South-East… 408 KP PRK 2006 35 182 0.29
8 Democrat… South-East… 408 KP PRK 2007 47 239 0.24
9 Democrat… South-East… 408 KP PRK 2008 57 293 0.2
10 Democrat… South-East… 408 KP PRK 2009 60 307 0.19
# ℹ 14 more rows
# ℹ 9 more variables: e_inc_100k <dbl>, e_inc_num <dbl>, e_mort_100k <dbl>,
# e_mort_exc_tbhiv_100k <dbl>, e_mort_exc_tbhiv_num <dbl>, e_mort_num <dbl>,
# e_mort_tbhiv_100k <dbl>, e_mort_tbhiv_num <dbl>, e_pop_num <dbl>5. 🔤 Text
cap <- "Week 45, 2025 | Source: World Health Organization Global Tuberculosis Report | Graphic by Andres Gonzalez"6. 📊 Plot
p <- who_tb_data %>%
ggplot(aes(x = e_inc_100k, y = e_mort_100k)) +
facet_wrap( ~ g_whoregion, scales = "free") +
geom_point(
data = ~ filter(., !country %in% highlighted_countries),
aes(
size = e_pop_num,
colour = g_whoregion,
fill = stage(g_whoregion, after_scale = alpha(colorspace::desaturate(fill, 0.4), 0.2))
),
shape = 21,
stroke = 0.2
) +
geom_point(
data = ~ filter(., country %in% highlighted_countries),
aes(size = e_pop_num, fill = g_whoregion),
shape = 21,
col = "white",
stroke = 0.15
) +
geom_text_repel(
data = . %>% filter(country %in% highlighted_countries),
aes(label = country),
fill = alpha("grey92", 0.5),
segment.color = "grey52",
size = 1.5,
family = "Roboto",
fontface = "bold",
seed = 10,
point.padding = 0.25,
min.segment.length = 0,
segment.size = 0.25
) +
scale_fill_manual(
values = c(
"#ef476f",
"#f78c6b",
"#ffd166",
"#06d6a0",
"#118ab2",
"#073b4c"
),
aesthetics = c("fill", "col")
) +
scale_size_area(
max_size = 20,
breaks = c(25, 100, 500, 1000) * 1e6,
labels = c("25M", "100M", "500M", "1B"),
guide = guide_circles(
override.aes = aes(colour = "grey45"),
title = "Population",
text_position = "ontop",
# style just this guide
theme = theme(
legend.title = element_text(hjust = 0.5),
legend.text = element_text(vjust = -0.5, size = 4)
)
)
) +
guides(fill = "none",
color = "none") +
labs(
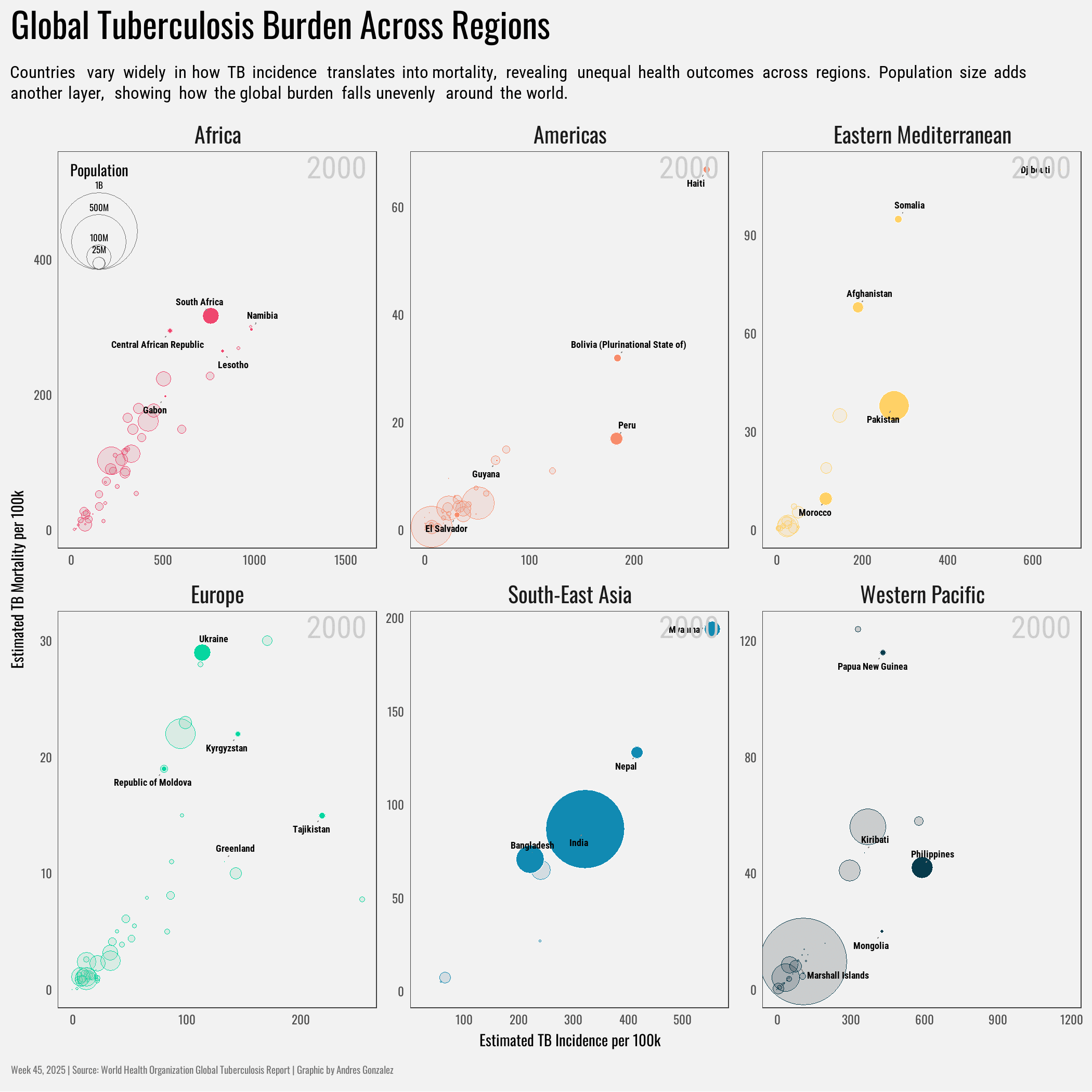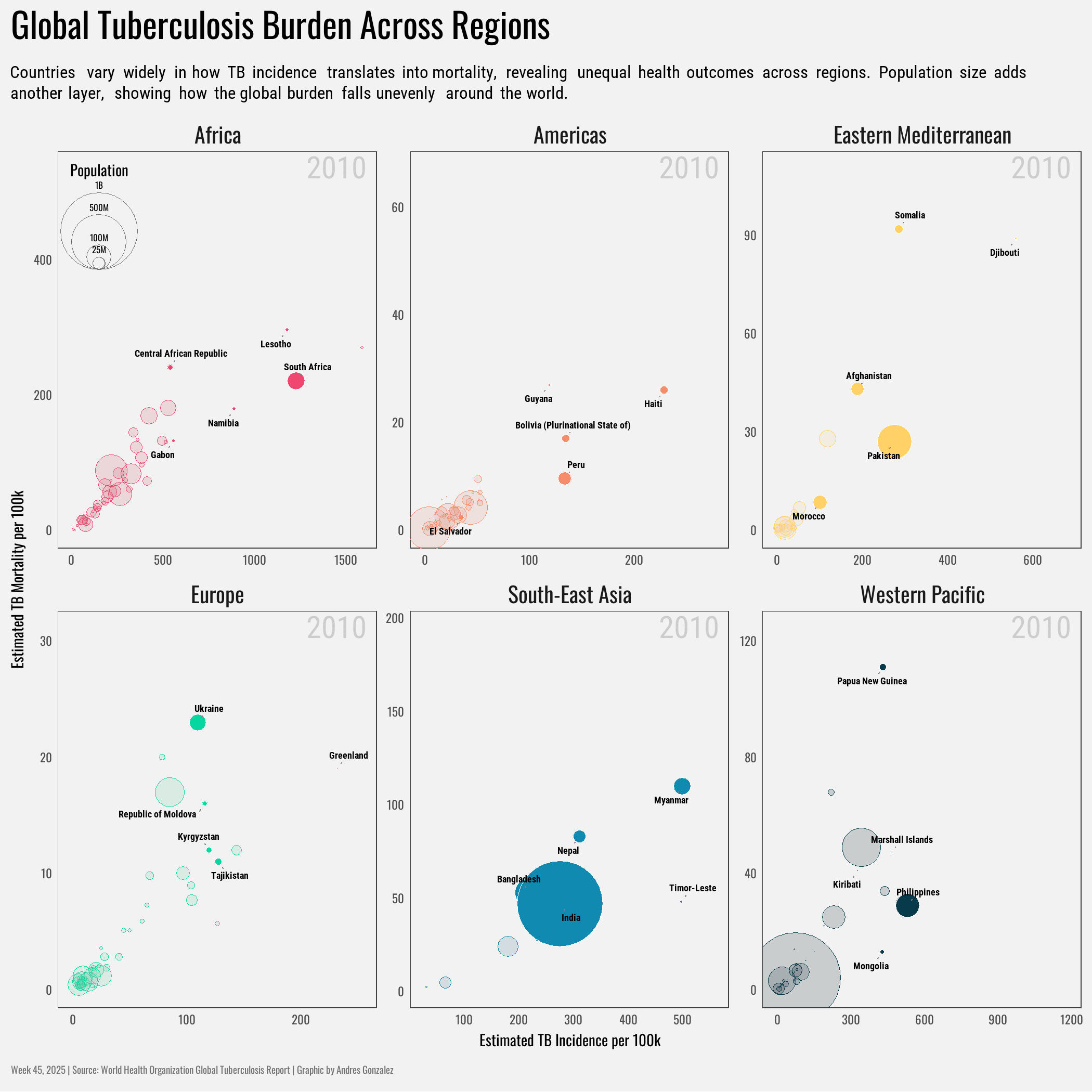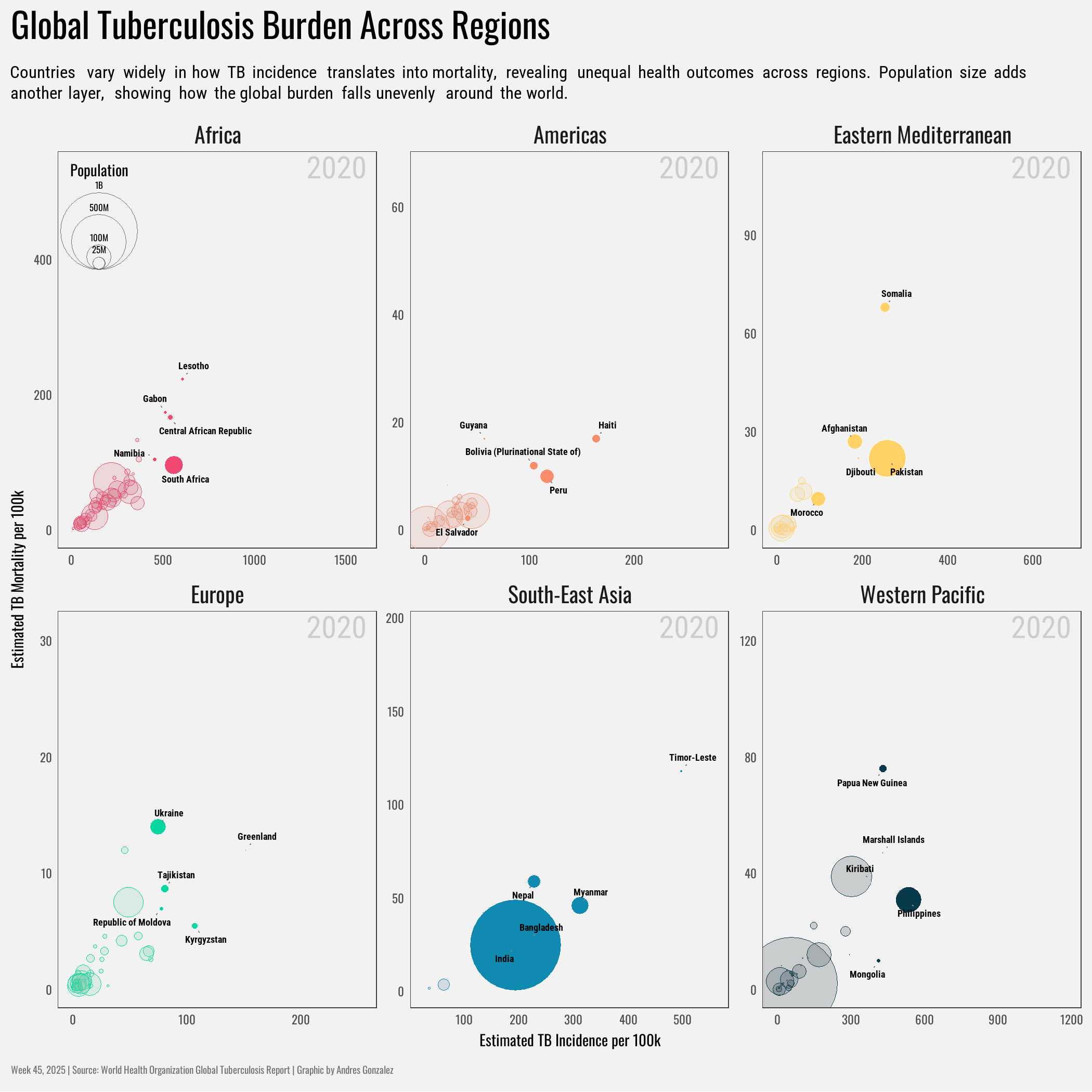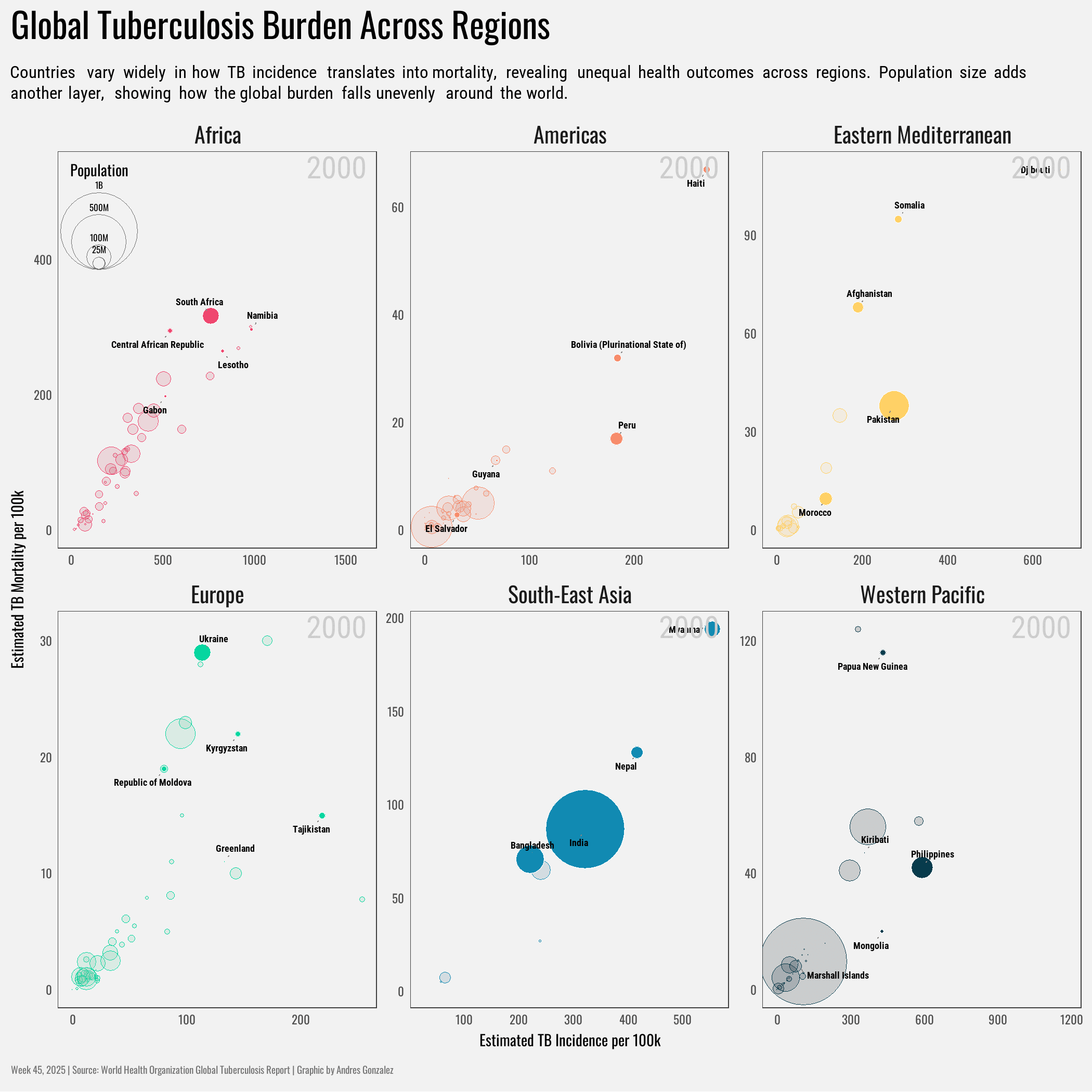
title = "Global Tuberculosis Burden Across Regions",
subtitle = "Countries vary widely in how TB incidence translates into mortality, revealing unequal health outcomes across regions. Population size adds another layer, showing how the global burden falls unevenly around the world.",
caption = cap,
x = "Estimated TB Incidence per 100k",
y = "Estimated TB Mortality per 100k"
) +
theme_minimal(base_size = 7, base_family = "Oswald") +
theme(
plot.title = element_text(
size = 16,
face = "bold",
margin = margin(b = 5)
),
plot.title.position = "plot",
plot.subtitle = element_textbox_simple(
family = "Roboto",
margin = margin(b = 7),
size = 8
),
plot.caption = element_text(
color = "grey45",
size = 4.5,
hjust = 0,
halign = 0,
margin = margin(t = 7)
),
plot.caption.position = "plot",
plot.margin = margin(5, 5, 5, 5),
panel.grid = element_blank(),
panel.background = element_rect(fill = "grey95", color = "grey25"),
plot.background = element_rect(fill = "grey95"),
strip.text = element_text(size = 10),
legend.position = c(0.04, 0.92)
) +
transition_states(year) +
geom_text(aes(label = scales::number(year, accuracy = 1, big.mark = ""),
x = Inf, y = Inf),
stat = "unique", color = "grey80", size = 5,
family = "Roboto",
vjust = 1.2, hjust = 1.2)
p <- animate(p, height = 7, width = 7, units = "in", res = 300, renderer = magick_renderer(loop = FALSE), duration = 15,
fps = 10,
end_pause = 36)
p1 <- who_tb_data %>%
ggplot(aes(x = e_inc_100k, y = e_mort_100k)) +
facet_wrap( ~ g_whoregion, scales = "free") +
geom_point(
data = ~ filter(., !country %in% highlighted_countries),
aes(
size = e_pop_num,
colour = g_whoregion,
fill = stage(g_whoregion, after_scale = alpha(colorspace::desaturate(fill, 0.4), 0.2))
),
shape = 21,
stroke = 0.2
) +
geom_point(
data = ~ filter(., country %in% highlighted_countries),
aes(size = e_pop_num, fill = g_whoregion),
shape = 21,
col = "white",
stroke = 0.15
) +
geom_text_repel(
data = . %>% filter(country %in% highlighted_countries),
aes(label = country),
fill = alpha("grey92", 0.5),
segment.color = "grey52",
size = 1.5,
family = "Roboto",
fontface = "bold",
seed = 10,
point.padding = 0.25,
min.segment.length = 0,
segment.size = 0.25
) +
scale_fill_manual(
values = c(
"#ef476f",
"#f78c6b",
"#ffd166",
"#06d6a0",
"#118ab2",
"#073b4c"
),
aesthetics = c("fill", "col")
) +
scale_size_area(
max_size = 20,
breaks = c(25, 100, 500, 1000) * 1e6,
labels = c("25M", "100M", "500M", "1B"),
guide = guide_circles(
override.aes = aes(colour = "grey45"),
title = "Population",
text_position = "ontop",
# style just this guide
theme = theme(
legend.title = element_text(hjust = 0.5),
legend.text = element_text(vjust = -0.5, size = 4)
)
)
) +
guides(fill = "none",
color = "none") +
labs(
title = "Global Tuberculosis Burden Across Regions",
subtitle = "Countries vary widely in how TB incidence translates into mortality, revealing unequal health outcomes across regions. Population size adds another layer, showing how the global burden falls unevenly around the world.",
caption = cap,
x = "Estimated TB Incidence per 100k",
y = "Estimated TB Mortality per 100k"
) +
theme_minimal(base_size = 7, base_family = "Oswald") +
theme(
plot.title = element_text(
size = 16,
face = "bold",
margin = margin(b = 5)
),
plot.title.position = "plot",
plot.subtitle = element_textbox_simple(
family = "Roboto",
margin = margin(b = 7),
size = 8
),
plot.caption = element_text(
color = "grey45",
size = 4.5,
hjust = 0,
halign = 0,
margin = margin(t = 7)
),
plot.caption.position = "plot",
plot.margin = margin(5, 5, 5, 5),
panel.grid = element_blank(),
panel.background = element_rect(fill = "grey95", color = "grey25"),
plot.background = element_rect(fill = "grey95"),
strip.text = element_text(size = 10),
legend.position = c(0.04, 0.92)
)7. 💾 Save
# Save Animation
anim_save("tt_45_2025.gif", p)
# Save the plot for TidyTuesday 2025, Week 45 with specified dimensions.
andresutils::save_plot(p1, type = "tidytuesday", year = 2025, week = 45, width = 9, height = 9)8. 🚀 GitHub Repository
TipExpand for GitHub Repo
The complete code for this analysis is available in tt_45_2025.qmd.
For the full repository, click here.
Citation
For attribution, please cite this work as:
Gonzalez, Andres. 2025. “Global Patterns in Tuberculosis Incidence
and Mortality.” November 12, 2025. https://andresgonzalezstats.com/visualization/TidyTuesday/2025/Week_45/tt_45_2025.html.